The RTMB package enables powerful and flexible statistical modelling with rich random effect structures using automatic differentiation (AD). However, its built-in support for probability distributions is limited to standard cases. RTMBdist fills this gap by providing a collection of non-standard, AD-compatible distributions, extending the range of models that can be implemented and estimated with RTMB. Most of the distributions implemented in RTMBdist allow for automatic simulation and residual calculation by RTMB.
The full list of distributions currently available is given in the List of distributions vignette. There are also a couple of Worked examples demonstrating how to use RTMBdist in practice.
Feel free to contribute or open an issue if you are missing a distribution!
Installation
You can install the released version from CRAN with:
install.packages("RTMBdist")or the development version from GitHub with:
remotes::install_github("janoleko/RTMBdist")Introductory example
Let’s do numerical maximum likelihood estimation with a gumbel distribution:
# simulate data
x <- rgumbel(100, location = 5, scale = 2)
# negative log-likelihood function
nll <- function(par) {
x <- OBS(x) # mark x as the response
loc <- par[1]; ADREPORT(loc)
scale <- exp(par[2]); ADREPORT(scale)
-sum(dgumbel(x, loc, scale, log = TRUE))
}
# RTMB AD object
obj <- MakeADFun(nll, c(5, log(2)), silent = TRUE)
# model fitting using AD gradient
opt <- nlminb(obj$par, obj$fn, obj$gr)
# model summary
summary(sdreport(obj))[3:4,]
#> Estimate Std. Error
#> loc 5.001543 0.2065935
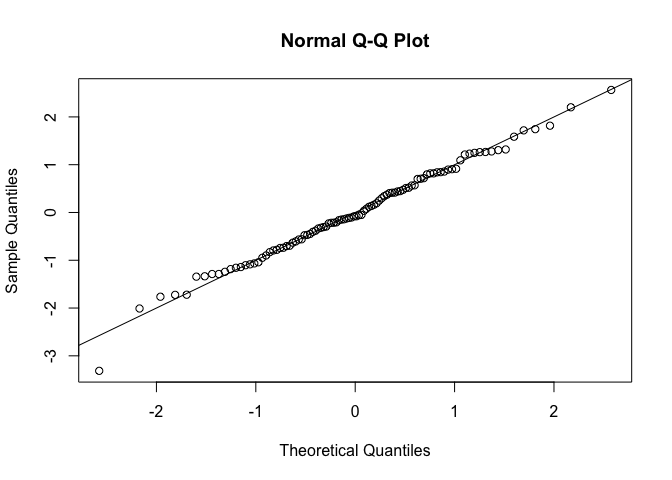
#> scale 1.960676 0.1502500Through the magic of RTMB, we can also immediately simulate new data from the fitted model and calculate residuals:
# simulate new data
x_new <- obj$simulate()$x
# calculate residuals
osa <- oneStepPredict(obj, method = "cdf", trace = FALSE)
qqnorm(osa$res); abline(0, 1)